- Rmarkdown Html Logo
- R Markdown Cheat Sheet
- Rmarkdown Html Create Table
- Rmarkdown Html Chunk
- Rmarkdown Html Page Width
- Rmarkdown Html Margin
The simplest way to write a quick report, mixing in a bit of R, is touse R Markdown, avariant of Markdowndeveloped by the folks at Rstudio.
You should first read the page about Markdown.
R Markdown is one of my favorite things about modern R. It offers an easy way to combine text, R code, and the results of R code in a single document. R Markdown will run the code and append the results to the doc. It will use the location of the.Rmd file as the working directory Code chunks Turn your report into an interactive Shiny document in 4 steps. Your report will rendered as a Shiny app, which means you must choose an html output format, like htmldocument. Including rmarkdown/html text in flexdashboard. Asked 4 years, 7 months ago. I have the following problem.
R Markdown
R Markdown is avariant of Markdownthat has embedded R code chunks, tobe used with knitr to make it easy tocreate reproducible web-based reports. The Markdown syntax has someenhancements (see theR Markdown page); for example,you can include LaTeX equations (seeEquations in R Markdown).
Here’s an example R Markdown document,and the html document it produces.
Code chunks
The key thing for us to focus on are the code chunks, which look like this:
In the midst of an otherwise plain Markdown document, you’ll have abit of R code that is initiated by a line like this:
After the code, there’ll be a line with just three backticks.
It’s usually best to give each code chunk a name, like simulate-dataand chunk-name above. The name is optional; if included, each codechunk needs a distinct name. The advantage of giving each chunk a nameis that it will be easier to understand where to look for errors,should they occur. Also, any figures that are created will be givennames based on the name of the code chunk that produced them. Asdiscussed in R Markdown: The DefinitiveGuide, it is bestfor the chunk labels to use just letters, numbers, and dashes, andno other special characters.
When you process the R Markdown document with knitr, each of the codechunks will be evaluated, and then the code and/or output will beinserted (unless you suppress one or both with chunk options, described below). Ifthe code produces a figure, that figure will be inserted.
An R Markdown document will have often have many code chunks. They areevaluated in order, in a single R session, and the state of thevarious variables in one code chunk are preserved in futurechunks. It’s as if you’d pulled out all of the R code as a single file(and you can dothat, using the purl command in knitr) and thensourceit into R.
Chunk options
The initial line in a code chunk may include various options. Forexample, echo=FALSE indicates that the code will not be shown in thefinal document (though any results/output would still be displayed).
You use results='hide' to hide the results/output (but here the codewould still be displayed).
You use include=FALSE to have the chunk evaluated Adobe air download free mac. , but neither thecode nor its output displayed.
If I’m writing a report for a collaborator, I’ll often useinclude=FALSE to suppress all of the code and largely just includefigures.
For figures, you’ll want to use options like fig.width andfig.height. For example:
Note that if include=FALSE, all of the code, results, and figureswill be suppressed. If include=TRUE and results='hide', the resultswill be hidden but figures will still be shown. To hide the figures,use fig.show='hide'.
There arelots of different possible “chunk options”.Each must be real R code, as R will be used to evaluate them. Soresults=hide is wrong; you need results='hide'.
Global chunk options
You may be inclined to use largely the same set of chunk optionsthroughout a document. But it would be a pain to retype those options in every chunk. Thus, you want to set someglobal chunk options at the top of your document.
For example, I might use include=FALSE or at least echo=FALSEglobally for a report to a scientific collaborator who wouldn’t wantto see all of the code. And I might want something like fig.width=12and fig.height=6 if I generally want those sizes for my figures.
I’d set such options by having an initial code chunk like this:
I snuck a few additional options in there: warning=FALSE andmessage=FALSE suppress any R warnings or messages from being included inthe final document, and fig.path='Figs/' makes it so the figurefiles get placed in the Figs subdirectory. (By default, they are notsaved at all.)
Note: the ending slash in Figs/ is important. If you usedfig.path='Figs' then the figures would go in the main directory butwith Figs as the initial part of their names.
The global chunk options become the defaults for the rest of thedocument. Then if you want a particular chunk to have a differentbehavior, for example, to have a different figure height, you’dspecify a different option within that chunk. For example:
In a report to a collaborator, I might use include=FALSE, echo=FALSEas a global option, and then use include=TRUE for the chunks thatproduce figures. Then the code would be suppressed throughout, and any outputwould be suppressed except in the figure chunks (where I usedinclude=TRUE), which would produce just the figures.
Technical aside: In setting the global chunk options withopts_chunk$set(), you’ll need to use knitr:: (or to have firstloaded the knitr package with library(knitr)). As we’ll discussbelow, we’ll use thermarkdown package to processthe document, first with knitr and then withpandoc, andrmarkdown::render() will use knitr::knit() but won’t loadthe knitr package.
Package options
In addition to the chunk options, there are alsopackage options,set with something like:
I was confused about this at first: I’d use opts_knit$set when Ireally wanted opts_chunk$set. knitr includes a lot of options; ifyou’re getting fancy you may need these package options, but initiallyyou’ll just be using the chunk options and, particularly, the globalchunk options defined via opts_chunk$set. So mostly ignoreopts_knit$set() in favor of opts_chunk$set().
In-line code
A key motivation for knitr isreproducible research:that our results are accompanied by the data and code needed toproduce them.
Thus, your report should never explicitly include numbers that arederived from the data. Don’t write “There are 168 individuals.”Rather, insert a bit of code that, whenevaluated, gives the number of individuals.
That’s the point of the in-line code. You’d write something like this:
Another example:
In R Markdown, in-line code is indicated with `r and `.The bit of R code between them is evaluated and the result inserted.
An important point: you need to be sure that these in-line bits ofcode aren’t split across lines in your document. Othewise you’ll justsee the raw code and not the result that you want.
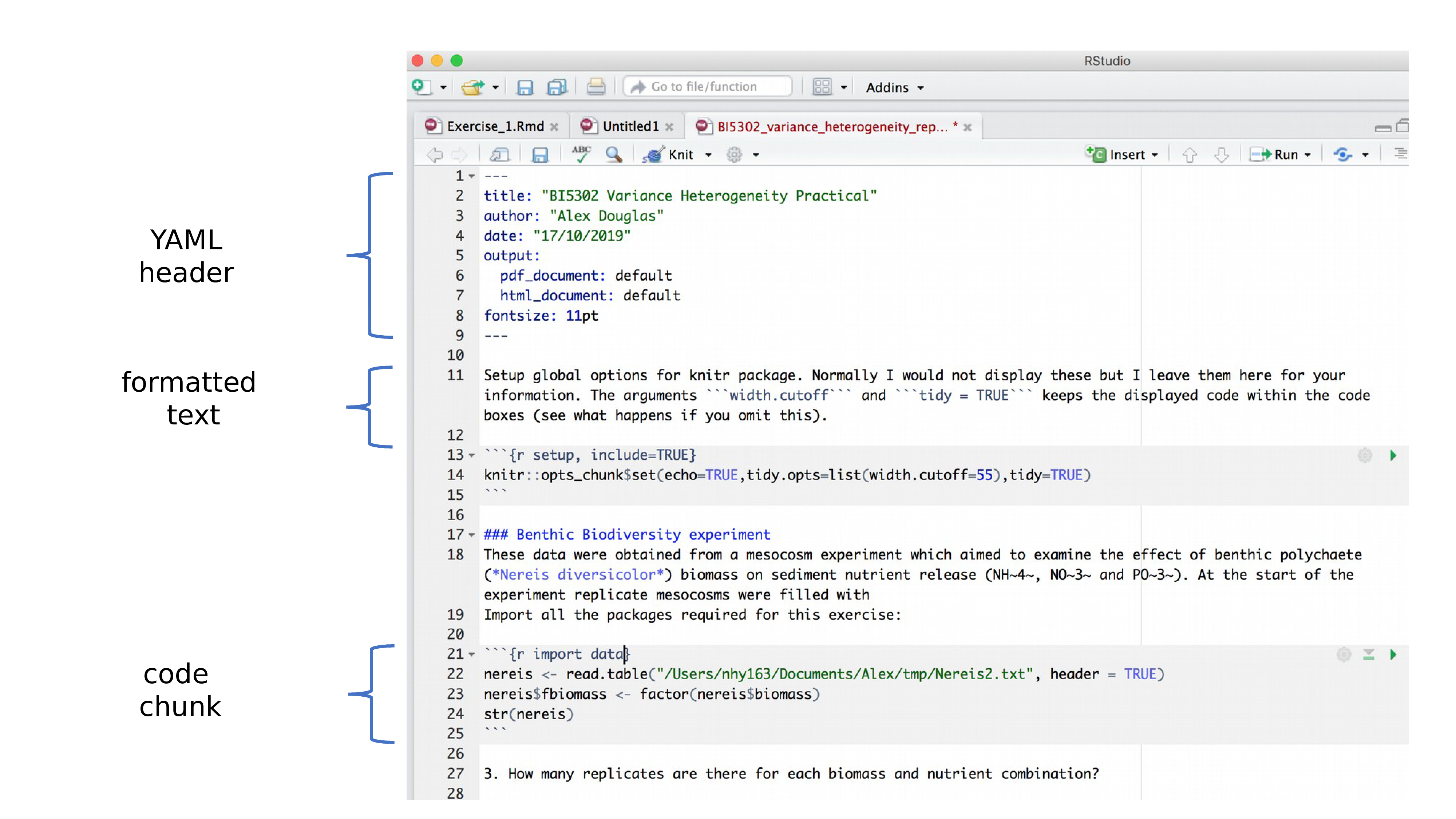
YAML header
Insert, at the top of your R Markdown document, a bit of text like thefollowing:
The final document will then contain a nicely formated title, alongwith the author name and date. Adobe after effects download free mac. You can include hyperlinks in there:
and even R code:
This is called the YAML header. YAML is asimple text-based format for specifying data, sort of likeJSON but more human-readable.
You can leave off the author and date if you want; you can leave offthe title, too. Actually, you don’t need to include any of this. Butoutput: html_document tells thermarkdown package to convertthe document to html. That’s the default, but you could also useoutput: pdf_document or even output: word_document, in which caseyour document will be converted to a PDF or Word .docx file,respectively.
Rounding
I’m very particular about the rounding of results, and you should be too. If I’ve estimateda correlation coefficient with 1000 data points, I don’t want to see0.9032738. I want 0.90.
You could use the R function round, like this: `r round(cor(x,y), 2)`But that would produce 0.9 instead of 0.90.
One solution is to use the sprintf function, like so:`r sprintf('%.2f', cor(x,y))`. That’s perfectly reasonable,right? Well, it is if you’re a C programmer.
But a problem arises if the value is -0.001. `r sprintf('%.2f', -0.001)`will produce -0.00. I don’t like that, nor doesHilary.
My solution to this problem is themyroundfunctionin my R/broman package.

At the start of my R Markdown document, I’d include:
And then later I could write `r myround(cor(x,y), 2)`and it would give 0.90 or 0.00 in the way that I want.
Converting R Markdown to html
Via RStudio
If you use RStudio, the simplest way toconvert an R Markdown document to html is to open the document withinRStudio. (And really, you probably want to create the document inRStudio: click File → New File → R Markdown.)When you open an R Markdown document in RStudio, you’ll seea “Knit HTML” button just above the document. (It’s a particularlycute little button, with a ball of yarn and a knitting needle.) Clickthat, and another window will open, and you’ll see knitr in action,executing each code chunk and each bit of in-line code, to compile the RMarkdown to a Markdown document. This will then be converted to html,with a preview of the result. (The resulting .html file will beplaced in the same directory as your .Rmd file.) You can click“Open in browser” to open the document in your web browser, or“Publish” to publish the document to the web (where it will beviewable by anyone).
Another a nice feature in RStudio: when you open an R Markdowndocument, you’ll see a little question mark button, with links to“Using R Markdown” and to a MarkdownQuick Reference. convenient “Markdown Quick Reference” document: acheat-sheet on the Markdown syntax. Like@StrictlyStat,I seem to visit theMarkdown website almostevery time I’m writing a Markdown document. If I used RStudio, I’dhave easier access to this information.
Apple macbook pro iso download. RStudio is especially useful when you’re first learning knitr and RMarkdown, as it’s easy to create and view the corresponding html file,and you have access to that Markdown Quick Reference.
Rmarkdown Html Logo
Via the command line (or GNU make)
To process an R Markdown document, you need thermarkdown package (which inturn will make use of theknitr package plus abunch of other packages), as well aspandoc.
To install the rmarkdown package, use install.packages(rmarkdown).
The simplest way to install pandoc is to just install theRStudio Desktop software,which includes pandoc, and then include pandoc without your PATH.On a Mac, you’d use:
In Windows, you’d include 'c:Program FilesRStudiobinpandoc' inyour Path system environment variable. (For example, seethis page,though it’s a bit ad-heavy.)
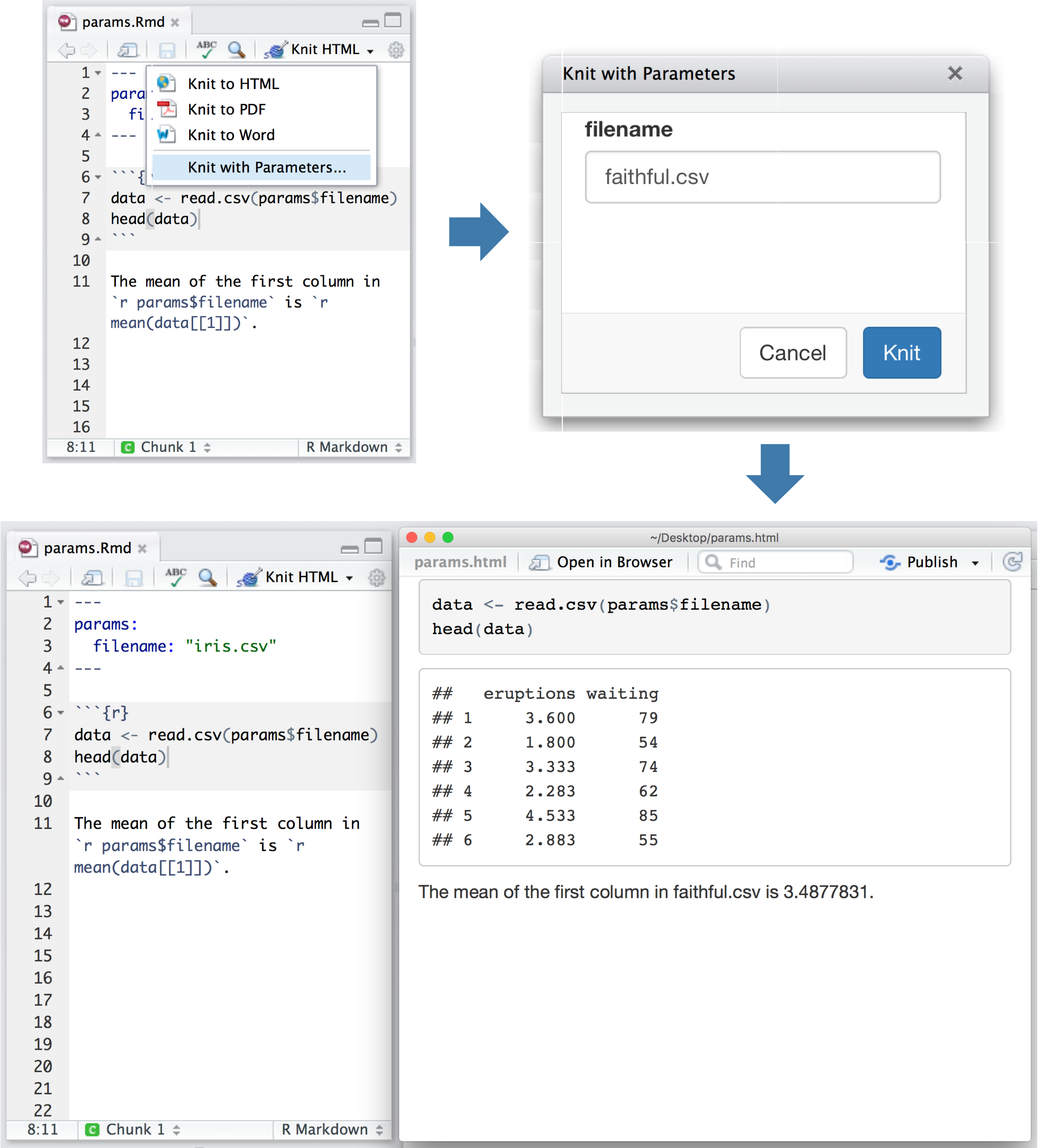
To convert your Markdown document to HTML, you’d then use
(Note that in Windows, it’s important to use double-quotes on theoutside and single-quotes inside, rather than the other way around.)
Rather than actually type that line, I include it within aGNU make file, likethis one.(Also see my minimal maketutorial.)
Up next
At this point, I’d recommend going off and playing with R Markdown fora while. Write your next report with R Markdown, even if it takes youa bit longer. Write it using RStudio, wherethe knitting process is easy and you have easy access to that“Markdown Quick Reference”.
Then, read a bit about figures and tables, mycomments on reproducibility, andperhaps about Knitr with AsciiDoc orKnitr with LaTeX.
In this tutorial, we will cover the R knitr package that is used to convertR Markdown into a rendered document (HTML, PDF, etc).
Learning Objectives
At the end of this activity, you will:
- Be able to produce (‘knit’) an HTML file from a R Markdown file.
- Know how to modify chunk options to change the output in your HTML file.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded onyour computer to complete this tutorial.
Install R Packages
- knitr:
install.packages('knitr') - rmarkdown:
install.packages('rmarkdown')
Share & Publish Results Directly from Your Code!
The knitr package allow us to:
- Publish & share preliminary results with collaborators.
- Create professional reports that document our workflow and results directlyfrom our code, reducing the risk of accidental copy and paste or transcription errors.
- Document our workflow to facilitate reproducibility.
- Efficiently change code outputs (figures, files) given changes in the data, methods, etc.
Publish from Rmd files with knitr
To complete this tutorial you need:
- The R
knitrpackage to complete this tutorial. If you need help installing packages, visit the R packages tutorial. - An R Markdown document that contains a YAML header, code chunks and markdownsegments. If you don't have an .Rmd file, visit the R Markdown tutorial to create one.
When To Knit: Knitting is a useful exercisethroughout your scientific workflow. It allows you to see what your outputslook like and also to test that your code runs without errors.The time required to knit depends on the length and complexity of the scriptand the size of your data.
How to Knit
To knit in RStudio, click the knit pull down button. You want to use the
knit HTML for this lesson.
R Markdown Cheat Sheet
When you click the Knit HTML button, a window will open in your console titled R Markdown. Thispane shows the knitting progress. The output (HTML in this case) file willautomatically be saved in the current working directory. If there is an errorin the code, an error message will appear with a line number in the R Consoleto help you diagnose the problem.
Data Tip: You can run knitr from the command promptusing: render(“input.Rmd”, “all”).
Activity: Knit Script
Rmarkdown Html Create Table
Knit the .Rmd file that you built inthe last tutorial.What does it look like?
View the Output

Rmarkdown Html Chunk
When knitting is complete, the new HTML file produced will automatically open.
Rmarkdown Html Page Width
Notice that information from the YAML header (title, author, date) is printedat the top of the HTML document. Then the HTML shows the text, code, andresults of the code that you included in the RMD document.
Data Institute Participants: Complete Week 2 Assignment
Rmarkdown Html Margin
- Read this week’s assignment closely.
- Be sure to carefully check your knitr output to make sure it is rendering theway you think it should!
- When you are complete, submit your .Rmd and .html files to the NEON Institute participants GitHub repository (NEONScience/DI-NEON-participants).
- The files will have automatically saved to your R working directory, you will need to transfer the files to the /participants/pre-institute3-rmd/ directory and submitted via a pull request.
